TLDR — Standard Model Bio (SMB) is building multimodal foundational models (FMs) for biopharma and owners of biomedical data (academic medical centers, biobanks, patients, etc). You can learn more about SMB in this Introducing Standard Model Biomedicine blog authored by SMB CEO/Co-Founder, Kevin Brown. Virtue led the SMB seed round.
Problem: Biopharma Moving towards Precision Medicine & AMCs Want To Participate, But Data Siloes Limit Scale
##Problem
Biopharma and AMCs are both moving towards precision medicine with multimodal data, but today that data is fragmented by institution, modality, and therapeutic area. Foundation models offer a shared backbone that lets biopharma and AMCs build together rather than in siloes.
Biopharma leveraging precision medicine to move beyond low-hanging fruit. Biopharma is being pushed to move towards precision medicine as they face an “innovation paradox”. A historic patent cliff over the next 5 years will lead to $200B+ in lost revenue while R&D productivity continues to decline (# of new drugs approved per $B in R&D halves every 9 years). To address the declining ROI of drug development, biopharma has been moving towards precision medicine (63% of approved drugs over the last decade). The first wave in oncology was successful (EGFR, KRAS, ALK, PD-1/PD-L1, etc) but future growth requires expansion to other data modalities and therapeutic areas.
AMCs have a history of participating in data partnerships, but engagement is limited and bespoke today. Over the last 10+ years, partnerships between AMCs and biopharma have shown the initial value of leveraging data to drive drug development. Regeneron and Geisenger partnered in 2014 on a DNA sequencing collaboration. Prometheus spun-off from Cedars in 2016 before being acquired by Merck for $11B. In 2016, Vanderbilt was selected by the NIH to lead the Data and Research Support Center for the Precision Medicine Initiative Cohort Program (now ‘All of Us’) and launched NashBio two years later. Culmination Bio launched out of Intermountain in 2023 to develop a disease-independent patient data intelligence platform. Work from industry, including Roche’s work with Flatiron (community oncology) and FMI (focused on AMCs), nference’s work with the Mayo Clinic platform, Tempus’ rich molecular data, and others, has also shown the value of these partnerships. Despite these examples of success, we need to scale these efforts across more data types and more therapeutic areas.
Data siloes limit performance: We still don’t have UK Biobank scale data networks in the US. Some of this is due to the natural siloes in biomedicine. Cancer care is divided into multiple sub-specialties, researchers study AI models for image segmentation in thoracic cancers while another may build models for genomic sequences in epilepsy, and biopharma has deep expertise in specific indications. For biomedicine to hit its scaling moment, we need multimodal, cross-therapeutic, and inter-organizational data.
Multimodal FMs can help solve some of these challenges for both AMCs and Biopharma, but talent for building biomedical foundational model infra is scarce. FMs can deliver better predictive accuracy, reliability across sites and adaptability across tasks and applications. These advantages allow the same backbone to drive increased accuracy and performance with multiple data types across multiple institutions with a variety of use cases. However, few people or teams have expertise in training foundation models at scale, experience with different data modalities (imaging, ‘omics, radiology, EMR, etc), AND have the biopharma domain expertise to understand what problems to tackle or questions to answer.
Enter Standard Model Bio
##Standard Model Bio
Standard Model Bio builds multimodal foundation models (’omics, radiology, pathology, EHRs, etc) for biopharma and owners of biomedical data (academic medical centers, biobanks, patients, etc). SMB’s mission is to accelerate biomedical AI by serving as its quiet backbone, providing customers and partners a compressed representation of human biomedical data via API or a fine-tunable model.
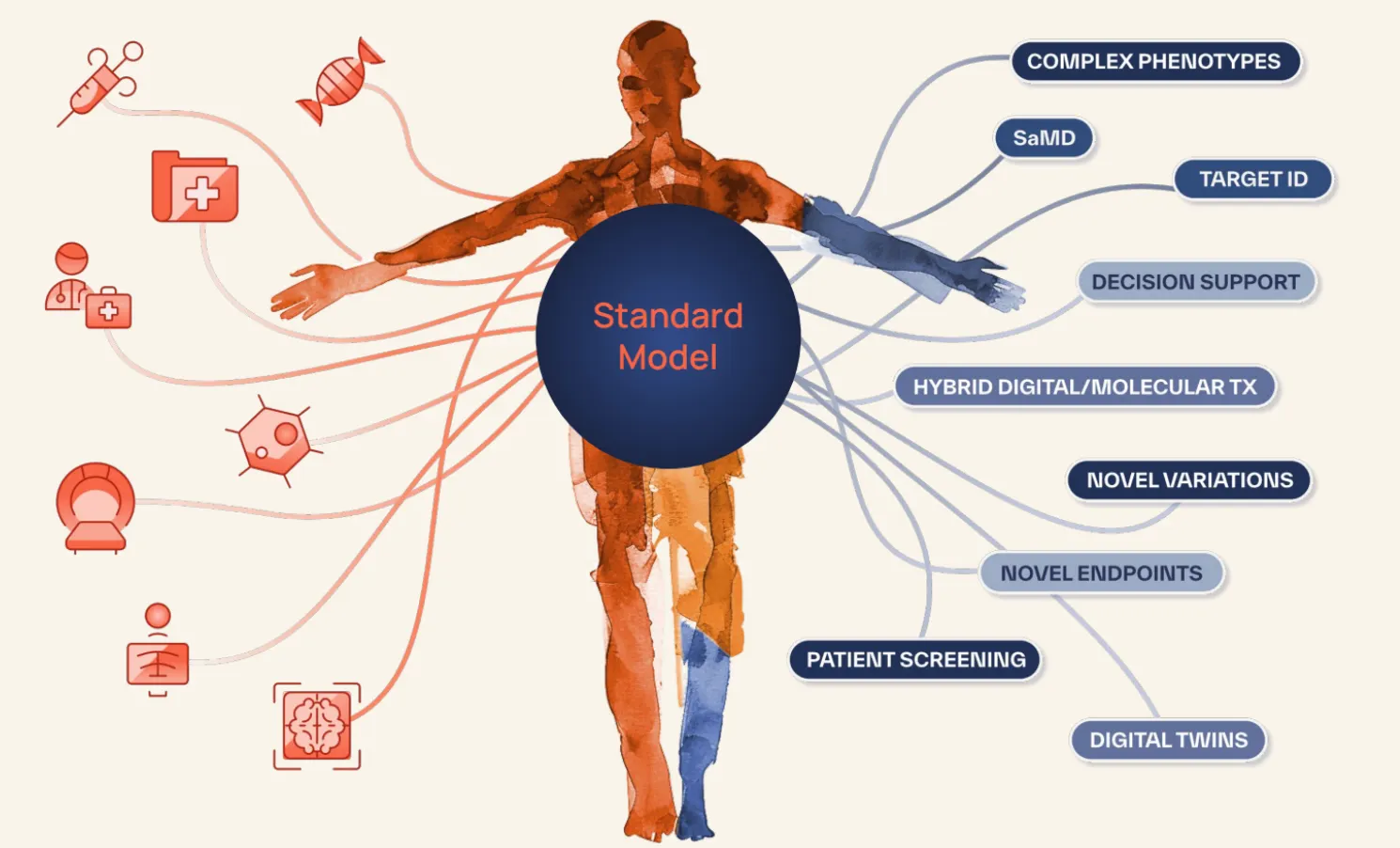
Downstream domain experts in biopharma or at health systems tune this model toward a variety of tasks, such as:
- Clinical trial recruitment
- Line-of-therapy transfer prediction for commercial arms of biopharma
- Predictive models for treatment selection (and equivalently companion-diagnostics for clinical trial acceleration)
- Digital twins via time-to-progression prognostic models
- Patient data validation for real-world evidence companies
- Image segmentation models for surgical preparation
SMB does not focus on protein/molecular design or chemistry. While we are excited about the advances in AI for drug discovery, the bottlenecks ahead will be testing these newly discovered therapies in the clinic and matching them to the right patients in clinical practice. Rather than try to predict the impact of cell perturbations in complex, 3-D cellular environments, SMB wants to answer a fundamental question by looking at patient outcomes and working backwards: What is the likely course & outcome of disease for this individual patient?
Team: Highly Concentrated Density Across Bio-Data-AI
##Team
The SMB team is uniquely positioned to build this future of biomedical AI. They have brought together the diverse expertise across different domains and data types required to build highly performant multimodal foundational models, and have been building over the last year to take theory into practice and demonstrate improved performance in the real-world.

Kevin Brown (CEO): Kevin previously led oncology foundation model development at Bristol Myers Squibb and federated learning R&D at Siemens Healthineers. His previous research spans state-of-the-art medical imaging foundation models, and the use of LLM's for clinical outcome prediction.
Zekai Chen (Multimodal/Time-series ML): Zekai previously built foundation models at JPMorganChase and Bristol Myers Squibb, and his previous research on graph learning, adaptive sharing, and drift-aware Transformers provide the basis of SMB’s foundation-model stack.
Arda Pekis (Medical imaging & VLMs): Arda focuses on high-resolution medical vision, from 3-D breast MRI segmentation validated across institutions to visualization tools that support surgical planning.
Shaun Porwal (Clinical utility & stats tooling): Shaun built stats/ML pipelines at Memorial Sloan Kettering (MSK) and open-sourced the dcurves library that biostatisticians now use to turn raw model scores into Decision Curve Analysis (DCA) plots (>32 k downloads).
David Laub (Genomics): David is a UC San Diego bioinformatics PhD who specializes in turning raw variant data into model-ready outputs, including his work on EUGENe, a FAIR deep-learning toolkit for regulatory genomics, and GenVarLoader, a memory-mapped data loader that massively accelerates personalized-genome training/inference.
David Laprade (Distributed Systems at Scale): David is a biologist and former software engineer at Stripe, where he invented a distributed computing algorithm that is the critical path of a system that moves >$1B/month across Stripe’s system.
Irsyad Adam (Multi-modal Graphs): Irsyad has previously built multi-modal AI pipelines for Genentech and AbbVie, and has created multi-modal graph deep learning pipelines and KG-LLM architectures for diagnosis, biomarker selection, and therapeutics.
Erik Reinertsen (Product & Partnerships): Erik previously built and led data science and engineering at Prometheus Biosciences through IPO to $10.8B acquisition by Merck, and is working to unlock similar value from real-world data for other academic medical centers and their biopharma partners.
Tech: Demonstrating Model Capabilities Across Diverse Data Types
##Tech
Building the Standard Model for biomedical AI requires data across several scales, from molecular to proteome-scale networks and tissue-level models, extending to whole-organ imaging, longitudinal EHRs, and patient outcomes.
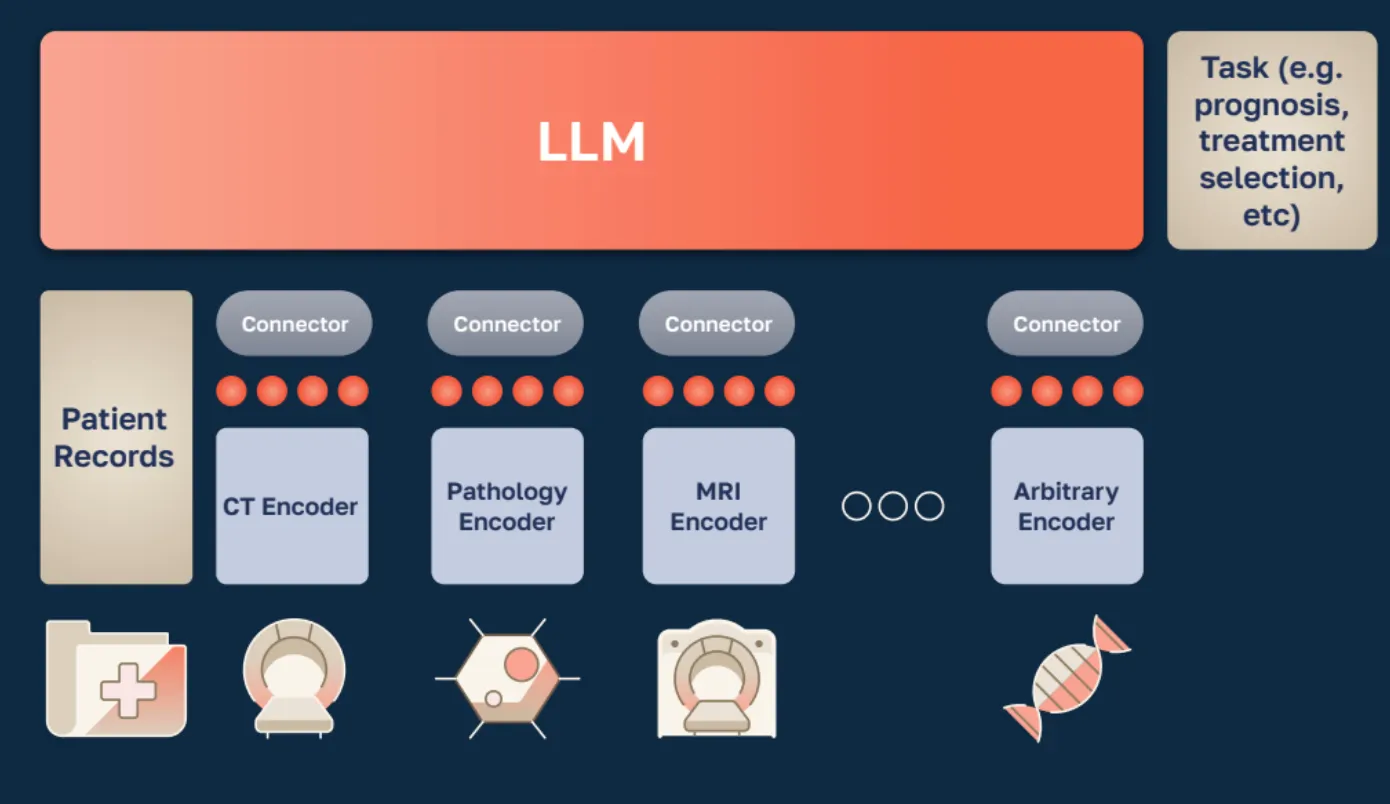
SMB just released three papers to showcase novel insights and core pieces that make up this holistic Standard Model.
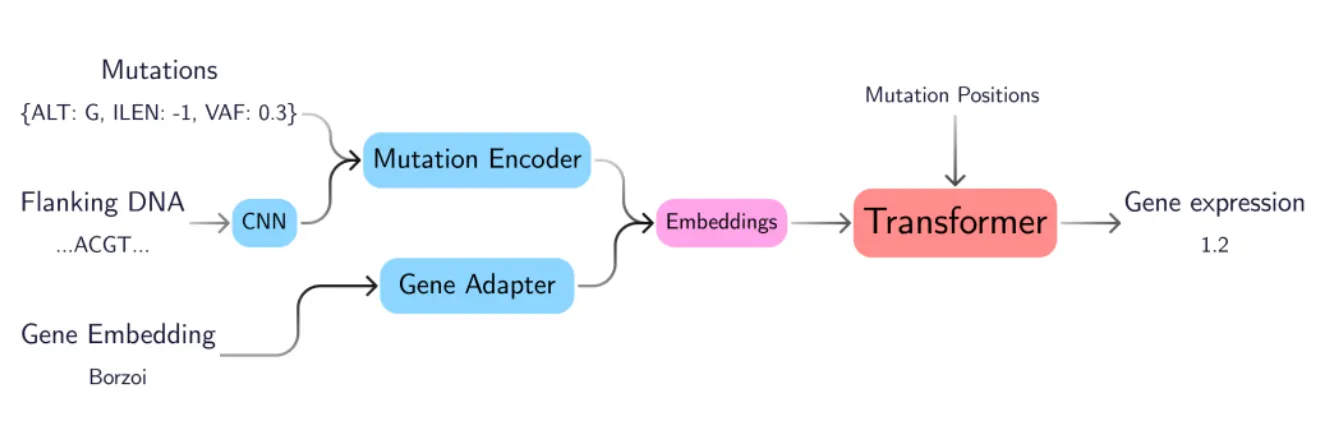
GenVarFormer: Predicting Gene Expression From Long-Range Mutations in Cancer
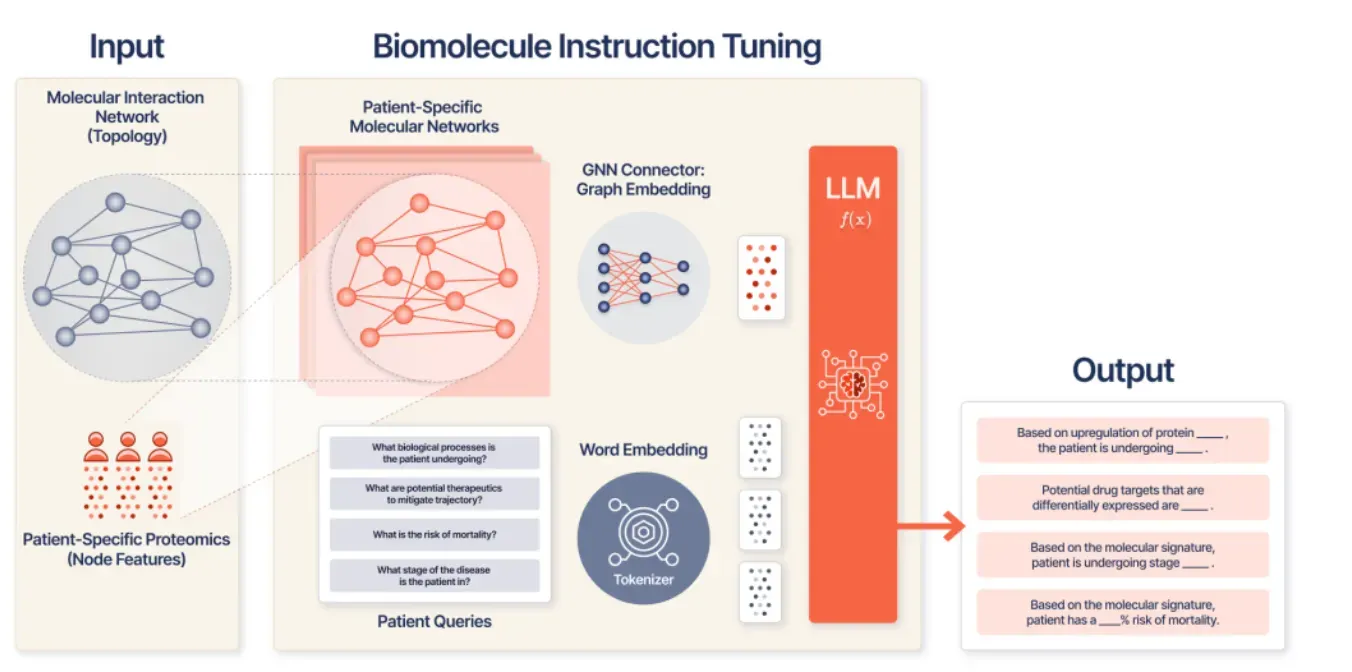
Patient-Specific Biomolecular Instruction Tuning of Graph-LLMs
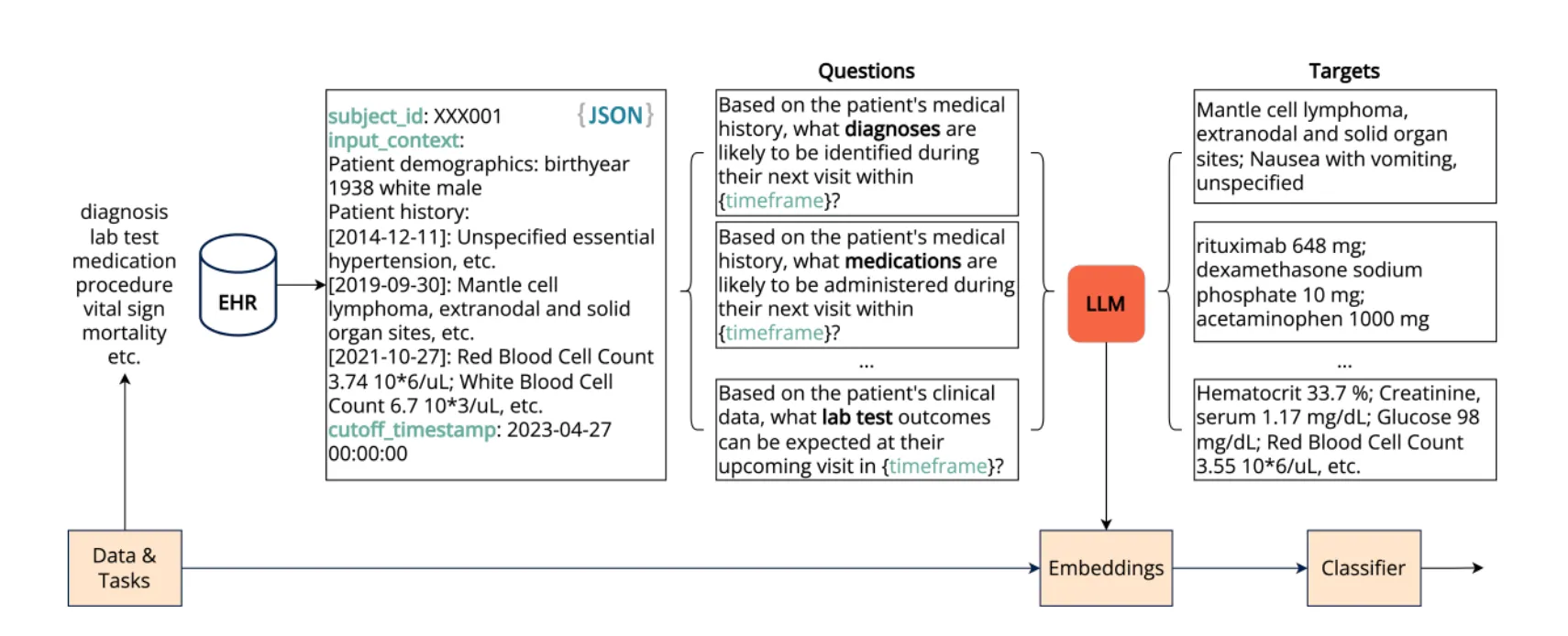
Building the EHR Foundation Model via Next Event Prediction (NEP)
These three papers represent SMB’s progress on three fundamental parts of the stack: molecular (WGS), cellular (proteomic networks + language), and whole-patient (longitudinal EHR). Together with SMB’s previous work on Advancing High Resolution Vision-Language Models in Biomedicine, which shows how their model improved upon the biology-specific large language and vision model LLaVA-Med by 10%, SMB has now produced SOTA work at nearly every scale of human biology.
Fundamentally, we have been impressed how (across multiple modalities and techniques) SMB takes it one step further to translate research and theory to clinical utility.
Others have built graph networks to predict protein-protein interactions (PPIs) to perturb networks and understand what genes/proteins are important. With KRONOS in Patient-Specific Biomolecular Instruction Tuning of Graph-LLMs, SMB instruction-tunes a graph-LLM over proteomic networks, enabling clinically relevant Q&A tasks such as relapse or stage prediction.
With GenVarFormer: Predicting Gene Expression From Long-Range Mutations in Cancer, SMB demonstrates similar capabilities. Not only did SMB demonstrate the technical architecture to handle non-coding drivers (rare, sparse, far from target genes), but this research connects mutations to gene expression to patient trajectories using patient embeddings that can be useful features for prognosis, subtype stratification, and drug response.
In its paper Building the EHR Foundation Model via Next Event Prediction (NEP), SMB builds on previous research showing that EHR foundational models can be modeled as sequences of events / tokens. We know performance hinges on longer context handling, and we need real-world, longitudinal few-shot evaluation. NEP is not only trained on fewer labels, but understands temporal events in EHRs so models learn the sequence of care instead of co-occurrence alone.
These research papers are previews of what is to come and there is more under the hood with SMB’s customers today as they continue to work with data at a larger and larger scale.
Vision For An Integrated Future
##Vision
Biology and medicine are multimodal because patients are inherently multimodal. Clinicians and researchers alike look at various data types:
- Molecular-level: Whole-genome sequencing, transcriptomics, proteomics
- Cell-level: CRISPR perturbation screens, “virtual cell” representations
- Tissue-level: Digital pathology and spatial transcriptomics
- Organ-level: CT scans, ECGs, EEGs
- Patient-level: Longitudinal EMRs and human activity data from wearables
This data is disconnected today on a “vertical level” (within an individual patient) but also on a horizontal-level - across drug discovery, drug development, and care delivery in the real-world. To accomplish the true vision for precision medicine we need to integrate on both of these axes.
Study design should be informed by commercial realities, site selection should be optimized for both recruitment and post-approval patient populations, and static treatment protocols in RCTs should match the dynamic, algorithmic treatment patterns we will see in practice over the next 3-5 years.
SMB epitomizes one of our core beliefs when it comes to today’s real-world data landscape and AI in biopharma: The prior era of data aggregation, recycling and re-selling is over — you now must deliver immediate value to the data originators and owners for the right and privilege to then leverage this data for novel downstream use cases.
The old data broker / data licensing model in biopharma was historically built on claims data from payers. FMs change this dynamic by providing broader opportunities for near-term operational and clinical value-add to data originators and owners. Unlike bespoke point solutions, FMs are extensible, can transfer across different modalities and patient types, and can actually surpass performance thresholds needed for real clinical impact, not just incremental utility for rare cases.
Next Steps
##Next Steps
Standard Model Bio (SMB) is always looking for people to join in building the future of biomedicine.
- If you are a biopharma company that wants to partner or a data source that wants to drive value from your data, reach out here.
- If you are both technical and biologically inclined and would like to join the SMB team, reach out here.
- If you would just like to chat about foundation models in biology then reach out here!
To stay updated, follow SMB on LinkedIn and X, and subscribe to their Substack.
The foundation is being laid for the future of biomedical research and drug development, one datapoint at a time.
